Fluid Mechanics: Navier-Stokes: 1D-0D Visible Human Example¶
Section author: David Ladd
Introduction¶
Computational models can be used to gain mechanical insight into blood flowing in vessels under healthy and pathophysiological states. However, constructing full subject-specific CFD models of the entire arterial and/or venous vasculature is currently considered impractical, owing to: (1) the time and resources required to identify, segment, and constrain a model of the billions of vessels in a human body and (2) the computational cost such a model would incur.
As a result, the systemic context of a modeled system is often incorporated into definable models through boundary conditions and/or coupled models. These multiscale models restrict higher dimensional (3D/1D) models to a region of interest and rely on simpler, lower dimensional (0D), models to approximate physical behaviour outside the higher-dimensional domain. This involves coupling together of dependent fields (ie pressure and velocity/flow), material fields (eg fluid viscosity and wall compliance), and geometric fields (eg vessel diameter) at the interfaces between 3D, 1D, and/or 0D model domains.
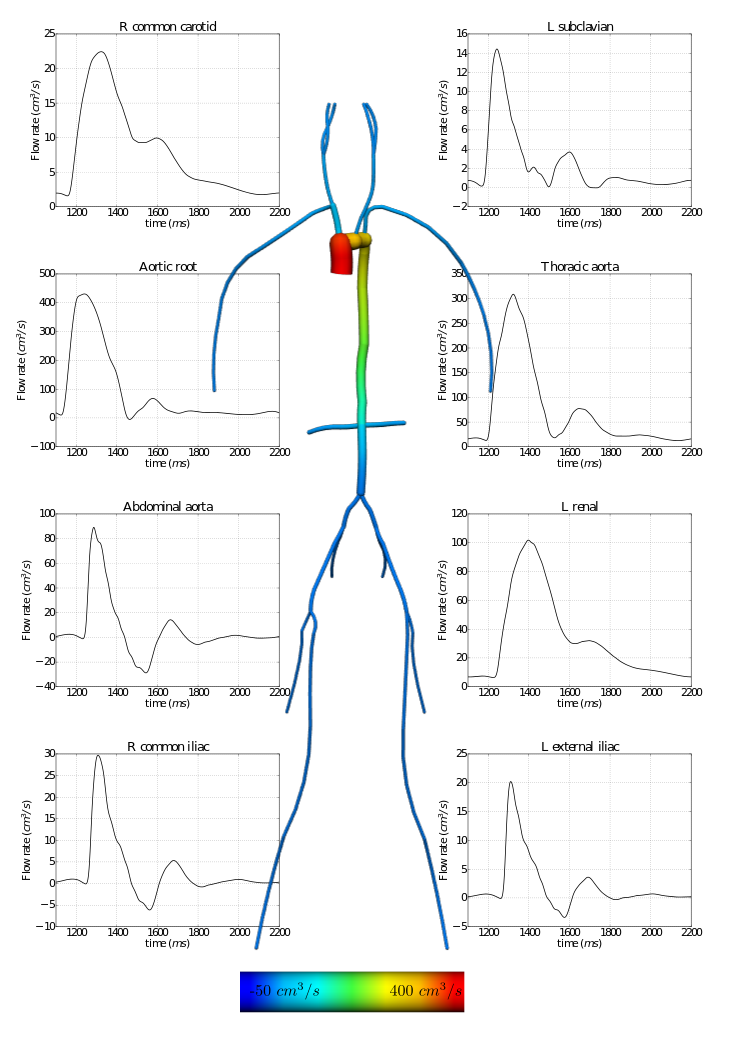
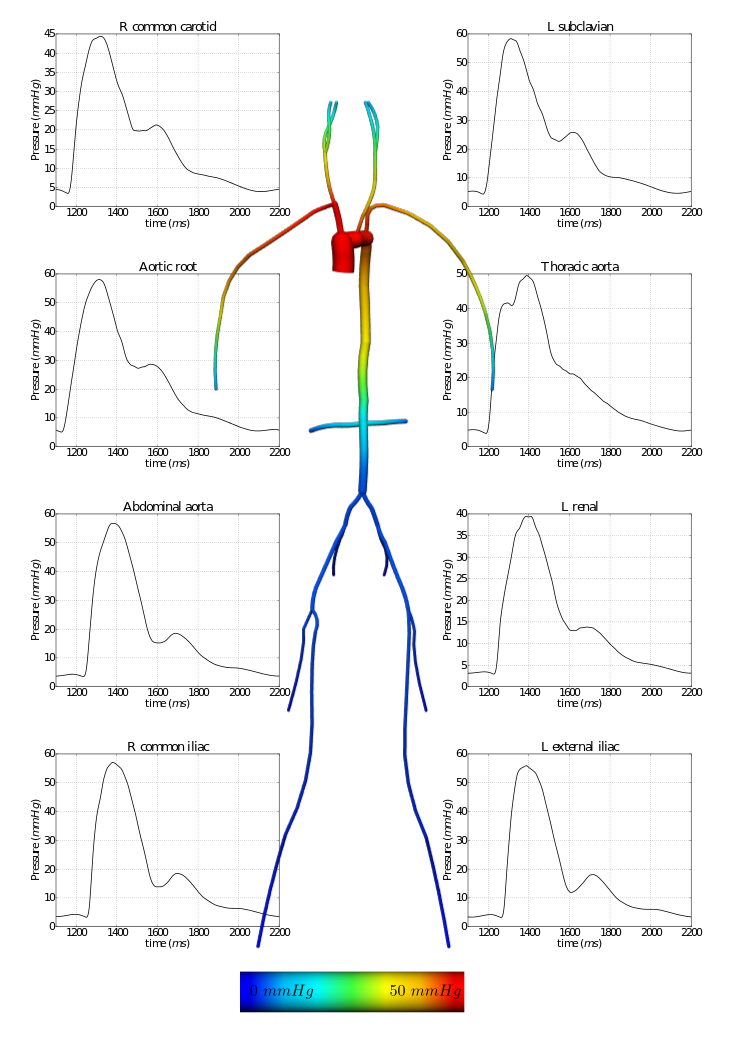
In the following example, a 1D network of 24 major arteries has been constructed from the male Visible Human dataset, as shown below. The resulting mesh contains 135 nodes on 67 quadratic line elements, as shown below.
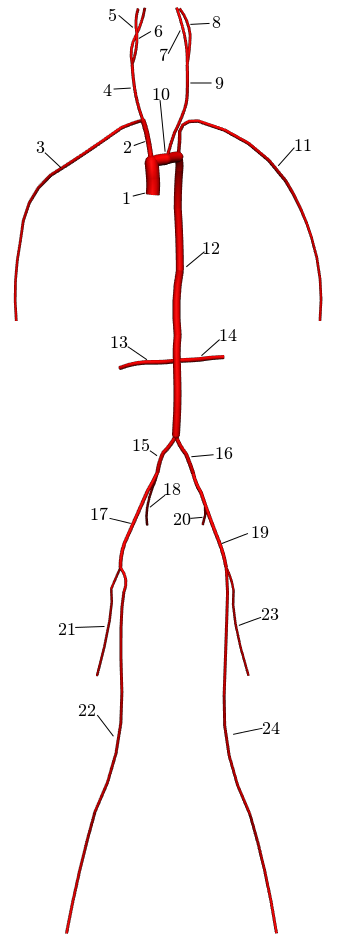
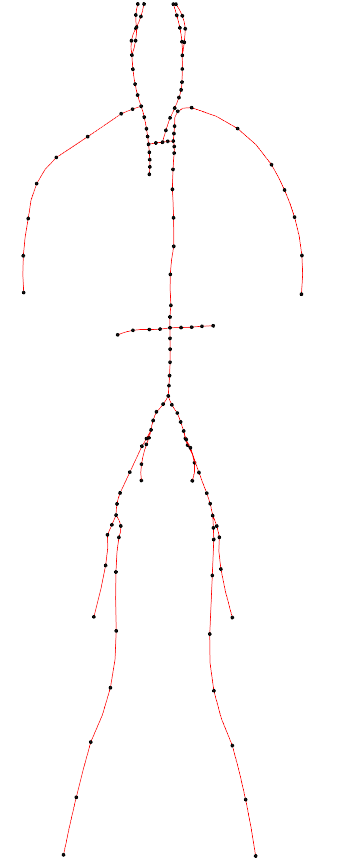
Figure: 1D arteries and mesh.
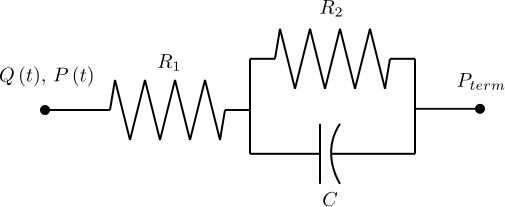
Simple 0D/lumped parameter RCR Windkessel models are coupled to the 1D model at the outlet boundaries. These models use the fluid-electric analog to provide an basic approximation of resistance to flow due to perfusing vascular beds (the R1 and R2 components) and downstream vessel compliance (the C component).
Methods¶
A flow waveform from published data is applied at the aortic root of the model and interpolated in time from tabulated data using cubic splines. Outlet boundary conditions are provided by the 0D solution.
OpenCMISS/CellML field mapping capabilities allow for the coupling of the 1D (OpenCMISS) and 0D (CellML) solvers:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 | #================================================================================================================================
# CellML Model Maps
#================================================================================================================================
if (RCRBoundaries):
#----------------------------------------------------------------------------------------------------------------------------
# Description
#----------------------------------------------------------------------------------------------------------------------------
# A CellML OD model is used to provide the impedance from the downstream vascular bed beyond the termination
# point of the 1D model. This is iteratively coupled with the the 1D solver. In the case of a simple resistance
# model, P=RQ, which is analogous to Ohm's law: V=IR. A variable map copies the guess for the FlowRate, Q at
# the boundary from the OpenCMISS Dependent Field to the CellML equation, which then returns presssure, P.
# The initial guess value for Q is taken from the previous time step or is 0 for t=0. In OpenCMISS this P value is
# then used to compute a new Area value based on the P-A relationship and the Riemann variable W_2, which gives a
# new value for Q until the values for Q and P converge within tolerance of the previous value.
#----------------------------------------------------------------------------------------------------------------------------
qCellMLComponent = 1
pCellMLComponent = 2
# Create the CellML environment
CellML = CMISS.CellML()
CellML.CreateStart(CellMLUserNumber,Region)
# Number of CellML models
CellMLModelIndex = [0]*(numberOfTerminalNodes+1)
# Windkessel Model
for terminalIdx in range (1,numberOfTerminalNodes+1):
nodeIdx = coupledNodeNumber[terminalIdx]
# Veins output
if (nodeIdx%2 == 0):
nodeDomain = Decomposition.NodeDomainGet(nodeIdx,meshComponentNumberSpace)
if nodeDomain == computationalNodeNumber:
CellMLModelIndex[terminalIdx] = CellML.ModelImport("./Input/CellMLModels/"+str(terminalIdx)+"/ModelHeart.cellml")
# known (to OpenCMISS) variables
CellML.VariableSetAsKnown(CellMLModelIndex[terminalIdx],"Heart/Qi")
CellML.VariableSetAsKnown(CellMLModelIndex[terminalIdx],"Heart/Po")
# to get from the CellML side
CellML.VariableSetAsWanted(CellMLModelIndex[terminalIdx],"Heart/Qo")
CellML.VariableSetAsWanted(CellMLModelIndex[terminalIdx],"Heart/Pi")
# Arteries outlet
else:
print('reading model: ' + "./Input/CellMLModels/"+str(terminalIdx)+"/ModelRCR.cellml")
nodeDomain = Decomposition.NodeDomainGet(nodeIdx,meshComponentNumberSpace)
if nodeDomain == computationalNodeNumber:
CellMLModelIndex[terminalIdx] = CellML.ModelImport("./Input/CellMLModels/"+str(terminalIdx)+"/ModelRCR.cellml")
# known (to OpenCMISS) variables
CellML.VariableSetAsKnown(CellMLModelIndex[terminalIdx],"Circuit/Qin")
# to get from the CellML side
CellML.VariableSetAsWanted(CellMLModelIndex[terminalIdx],"Circuit/Pout")
CellML.CreateFinish()
# Start the creation of CellML <--> OpenCMISS field maps
CellML.FieldMapsCreateStart()
# ModelIndex
for terminalIdx in range (1,numberOfTerminalNodes+1):
nodeIdx = coupledNodeNumber[terminalIdx]
# Veins output
if (nodeIdx%2 == 0):
nodeDomain = Decomposition.NodeDomainGet(nodeIdx,meshComponentNumberSpace)
if nodeDomain == computationalNodeNumber:
# Now we can set up the field variable component <--> CellML model variable mappings.
# Map the OpenCMISS boundary flow rate values --> CellML
# Q is component 1 of the DependentField
CellML.CreateFieldToCellMLMap(DependentFieldNavierStokes,CMISS.FieldVariableTypes.U,1,
CMISS.FieldParameterSetTypes.VALUES,CellMLModelIndex[terminalIdx],"Heart/Qi",CMISS.FieldParameterSetTypes.VALUES)
CellML.CreateFieldToCellMLMap(DependentFieldNavierStokes,CMISS.FieldVariableTypes.U2,1,
CMISS.FieldParameterSetTypes.VALUES,CellMLModelIndex[terminalIdx],"Heart/Po",CMISS.FieldParameterSetTypes.VALUES)
# Map the returned pressure values from CellML --> CMISS
# pCellML is component 2 of the Dependent field U1 variable
CellML.CreateCellMLToFieldMap(CellMLModelIndex[terminalIdx],"Heart/Qo",CMISS.FieldParameterSetTypes.VALUES,
DependentFieldNavierStokes,CMISS.FieldVariableTypes.U1,qCellMLComponent,CMISS.FieldParameterSetTypes.VALUES)
CellML.CreateCellMLToFieldMap(CellMLModelIndex[terminalIdx],"Heart/Pi",CMISS.FieldParameterSetTypes.VALUES,
DependentFieldNavierStokes,CMISS.FieldVariableTypes.U1,pCellMLComponent,CMISS.FieldParameterSetTypes.VALUES)
# Arteries output
else:
nodeDomain = Decomposition.NodeDomainGet(nodeIdx,meshComponentNumberSpace)
if nodeDomain == computationalNodeNumber:
# Now we can set up the field variable component <--> CellML model variable mappings.
# Map the OpenCMISS boundary flow rate values --> CellML
# Q is component 1 of the DependentField
CellML.CreateFieldToCellMLMap(DependentFieldNavierStokes,CMISS.FieldVariableTypes.U,1,
CMISS.FieldParameterSetTypes.VALUES,CellMLModelIndex[terminalIdx],"Circuit/Qin",CMISS.FieldParameterSetTypes.VALUES)
# Map the returned pressure values from CellML --> CMISS
# pCellML is component 1 of the Dependent field U1 variable
CellML.CreateCellMLToFieldMap(CellMLModelIndex[terminalIdx],"Circuit/Pout",CMISS.FieldParameterSetTypes.VALUES,
DependentFieldNavierStokes,CMISS.FieldVariableTypes.U1,pCellMLComponent,CMISS.FieldParameterSetTypes.VALUES)
# Finish the creation of CellML <--> OpenCMISS field maps
CellML.FieldMapsCreateFinish()
CellMLModelsField = CMISS.Field()
CellML.ModelsFieldCreateStart(CellMLModelsFieldUserNumber,CellMLModelsField)
CellML.ModelsFieldCreateFinish()
# Set the models field at boundary nodes
for terminalIdx in range (1,numberOfTerminalNodes+1):
nodeIdx = coupledNodeNumber[terminalIdx]
nodeDomain = Decomposition.NodeDomainGet(nodeIdx,meshComponentNumberSpace)
if nodeDomain == computationalNodeNumber:
CellMLModelsField.ParameterSetUpdateNode(CMISS.FieldVariableTypes.U,CMISS.FieldParameterSetTypes.VALUES,
versionIdx,derivIdx,nodeIdx,1,CellMLModelIndex[terminalIdx])
CellMLStateField = CMISS.Field()
CellML.StateFieldCreateStart(CellMLStateFieldUserNumber,CellMLStateField)
CellML.StateFieldCreateFinish()
CellMLParametersField = CMISS.Field()
CellML.ParametersFieldCreateStart(CellMLParametersFieldUserNumber,CellMLParametersField)
CellML.ParametersFieldCreateFinish()
CellMLIntermediateField = CMISS.Field()
CellML.IntermediateFieldCreateStart(CellMLIntermediateFieldUserNumber,CellMLIntermediateField)
CellML.IntermediateFieldCreateFinish()
# Finish the parameter update
DependentFieldNavierStokes.ParameterSetUpdateStart(CMISS.FieldVariableTypes.U1,CMISS.FieldParameterSetTypes.VALUES)
DependentFieldNavierStokes.ParameterSetUpdateFinish(CMISS.FieldVariableTypes.U1,CMISS.FieldParameterSetTypes.VALUES)
|
Snippet: OpenCMISS/CellML field mappings
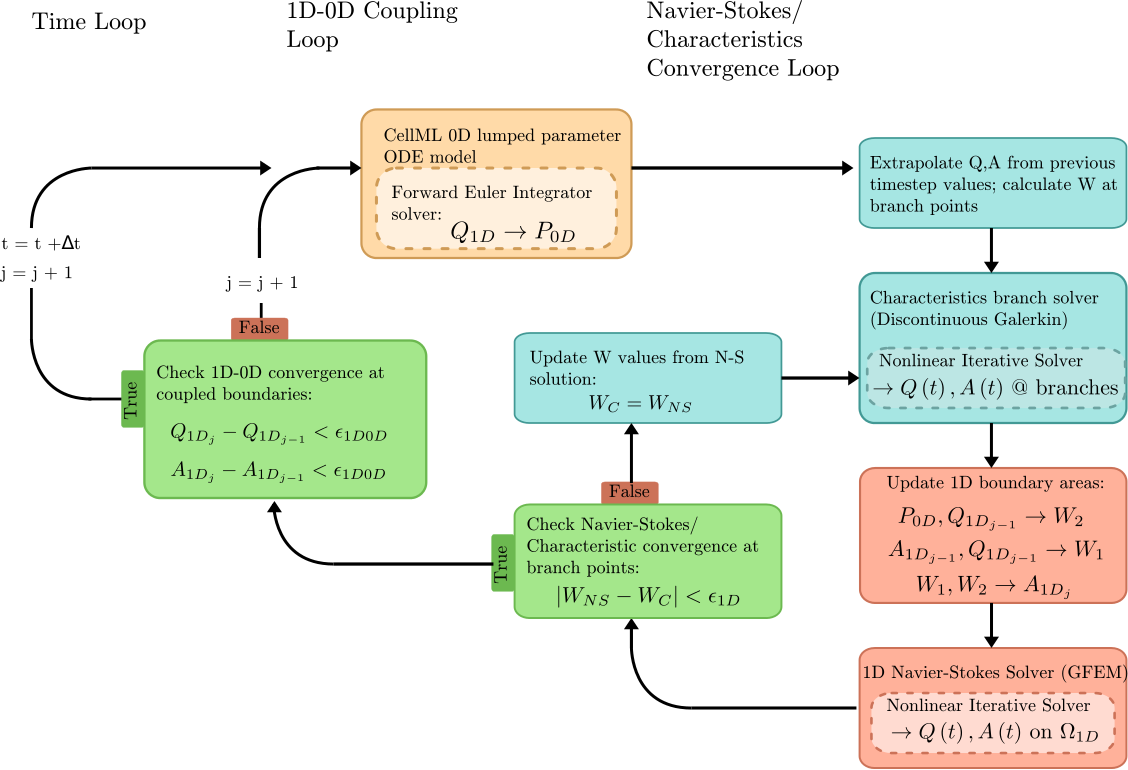
Flow rate (Q) from the 1D Navier-Stokes/Characteristic solver provides the forcing term for the 0D ODE circuit solver. Pressure (P) is returned from the CellML solver to provide constraints on the Riemann invariants of the 1D system, which translate to area boundary conditions for the 1D FEM solver. At each timestep, the 1D and 0D systems are iteratively coupled until the boundary values converge within a user-specified tolerance at the 1D-0D interfaces. This procedure is outlined in the figure below.